Intro
I am a Ph.D. candidate in the department of Biomedical Engineering, University of Melbourne. I am being advised by Dr. Vijay Rajagopal and Professor Thomas Boudier. I have also collaborated closely with Professor Eric Hanssen.
My research focuses on developing data-driven and large-scale computational methods for biological image analysis and healthcare problems. In particular, I make use of techniques in machine learning, deep learning, image processing, algorithms, and data analytics to solve massive-scale problems that stem from real-world data, with a special focus on biological/biomedical image analysis.
Since joining Cell Structure and MechanoBiology group, I have achieved several accomplishments including 1st place across Australia and 8th globally among more than top 200 institutes and research groups worldwide participating MIT ISBI 2012 Challenge on Neuronal Image segmentation. My conference paper presented in IEEE BIBM 2018 had been selected for special issue publication in BMC Medical Informatics and Decision Making among more than 500 papers published in conference proceedings.
Research
EM-stellar
Benchmarking deep learning for electron microscopy image segmentation
 |
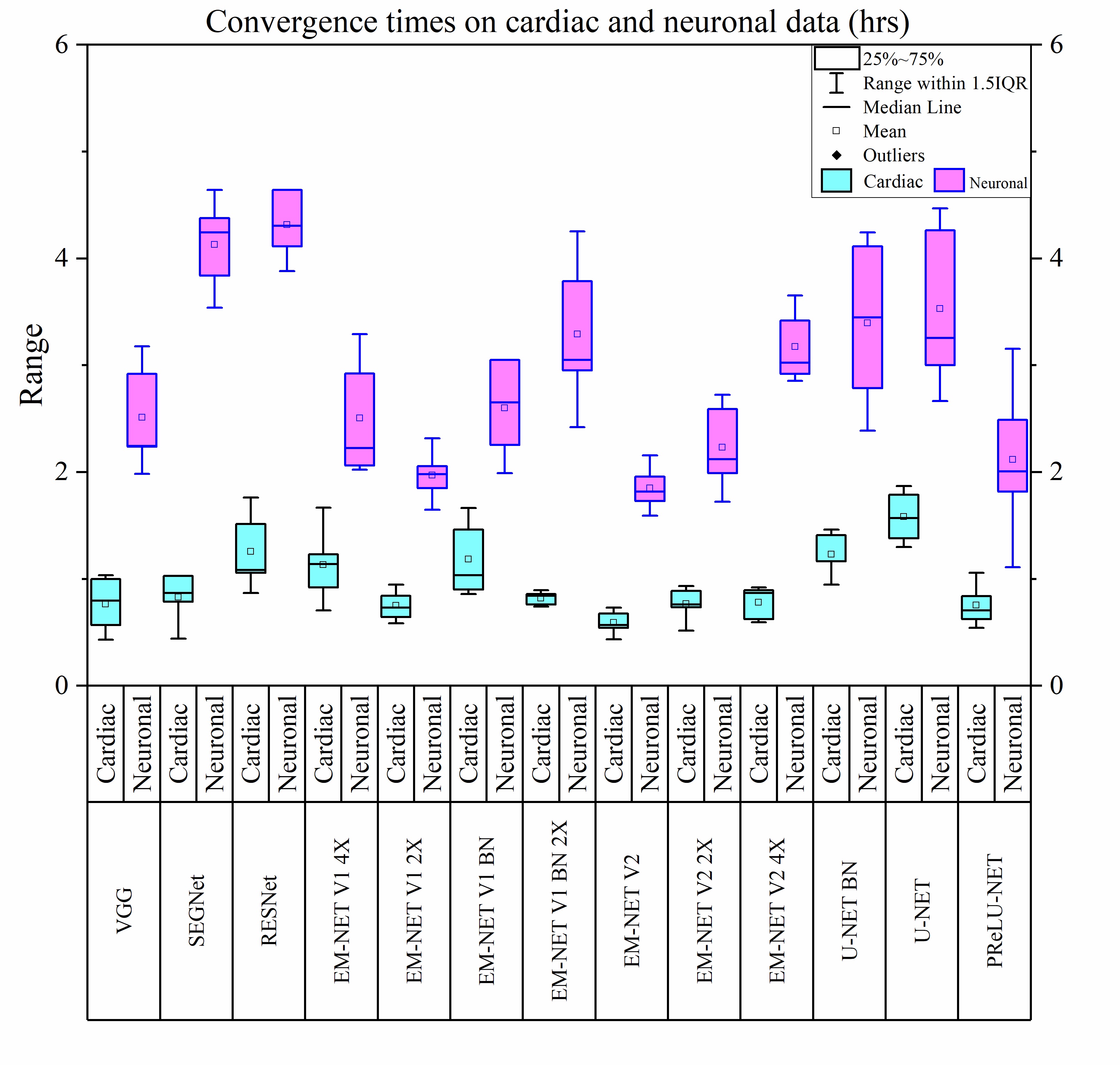 |
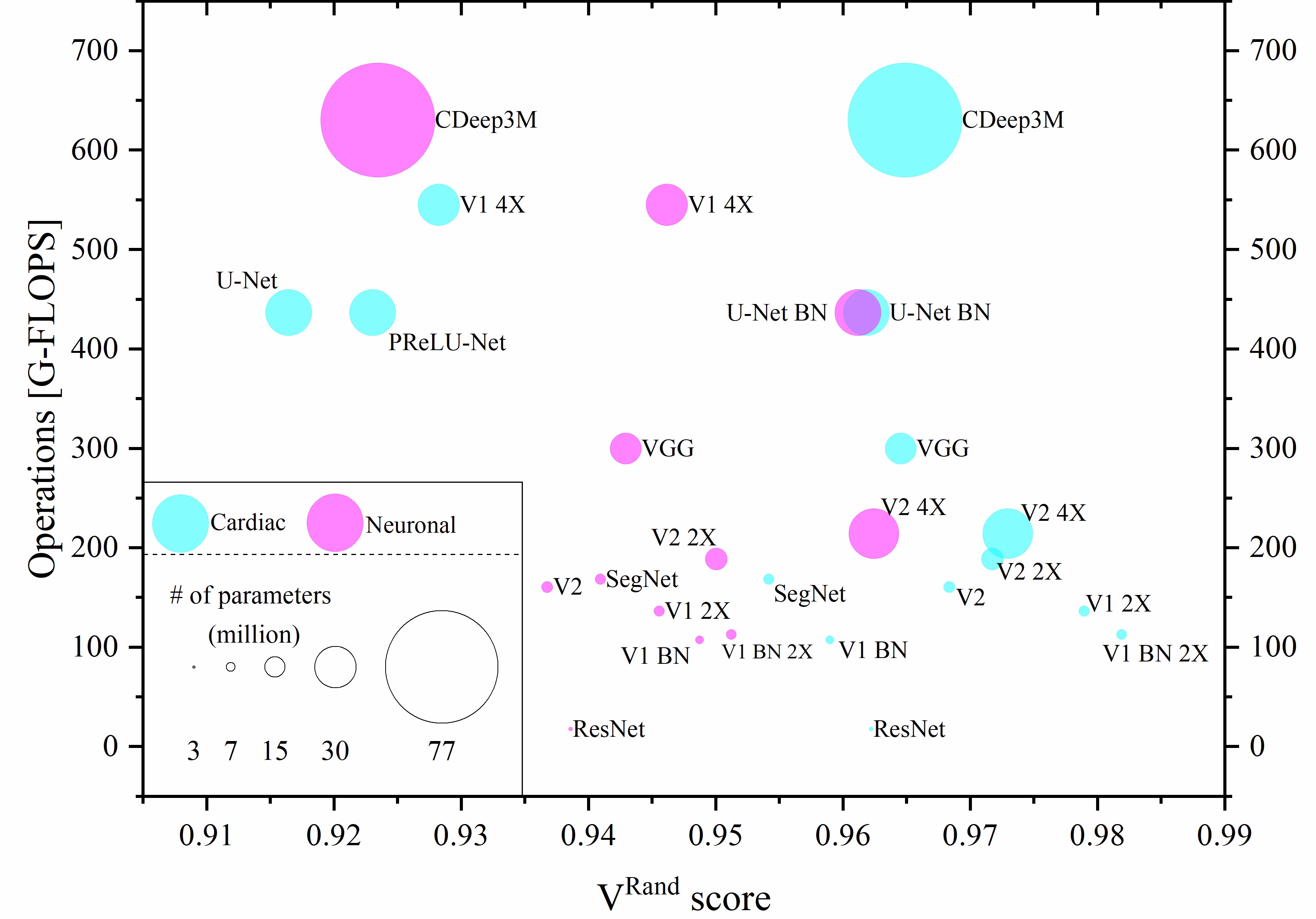
The inherent low contrast of electron microscopy (EM) datasets presents a significant challenge for rapid segmentation of cellular ultrastructures from EM data. This challenge is particularly prominent when working with high-resolution big-datasets that are now acquired using electron tomography and serial block-face imaging techniques. Deep learning (DL) methods offer an exciting opportunity to automate the segmentation process by learning from manual annotations of a small sample of EM data. While many DL methods are being rapidly adopted to segment EM data no benchmark analysis has been conducted on these methods to date. We present EM-stellar, a platform that is hosted on Google Colab that can be used to benchmark the performance of a range of state-of-the-art DL methods on user-provided datasets. Using EM-stellar we show that the performance of any DL method is dependent on the properties of the images being segmented. It also follows that no single DL method performs consistently across all performance evaluation metrics.
Related publications:
1 - Khadangi, Afshin, Thomas Boudier, and Vijay Rajagopal. "EM-stellar: benchmarking deep learning for electron microscopy image segmentation.
|
dEM0ise: dynamic deep learning for zero-noise EM data
 |
 |
Deep learning has been successful in a wide variety of computer vision applications including image segmentation, object classification, tracking, style transfer, colourisation and image denoising. Electron microscopy data such as FIB/SBF-SEM images provide high-throughput information on the sub-cellular level which assist biologists in deriving imperative insights to the cell function and organisation. However, these types of datasets are intrinsically noisy and sometimes need sophisticated preprocessing methods to enhance these images before other critical tasks such as object segmentation.
We have addressed all these barriers with dEM0ise, a web-based EM denoising hub for biologists. One of the essential aims of dEM0ise is to expand the database of EM images which will extend the capability of dEM0ise to the next level.
Related publications:
Coming soon!.
|
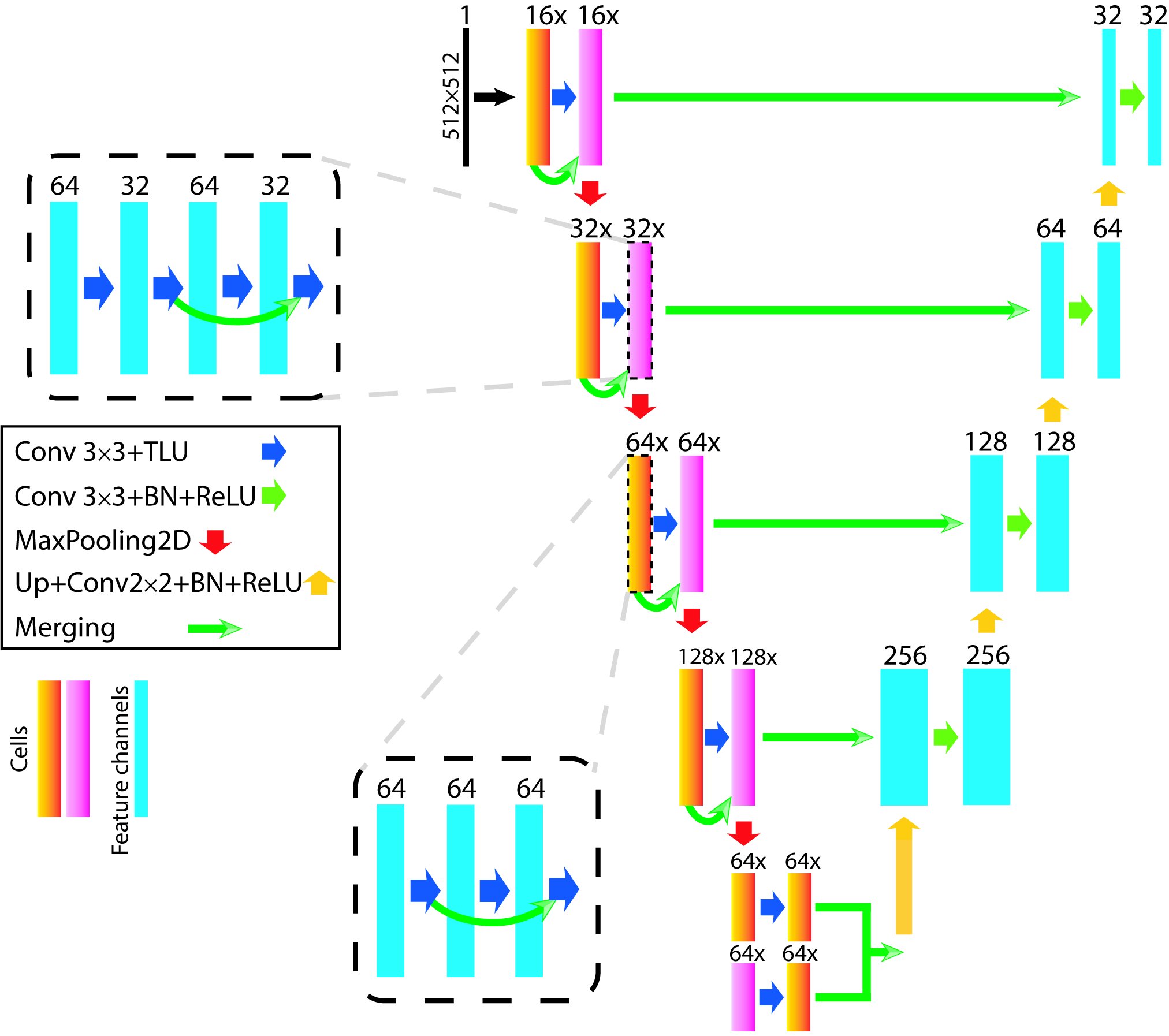
EM-net
Deep learning for electron microscopy image segmentation
 |
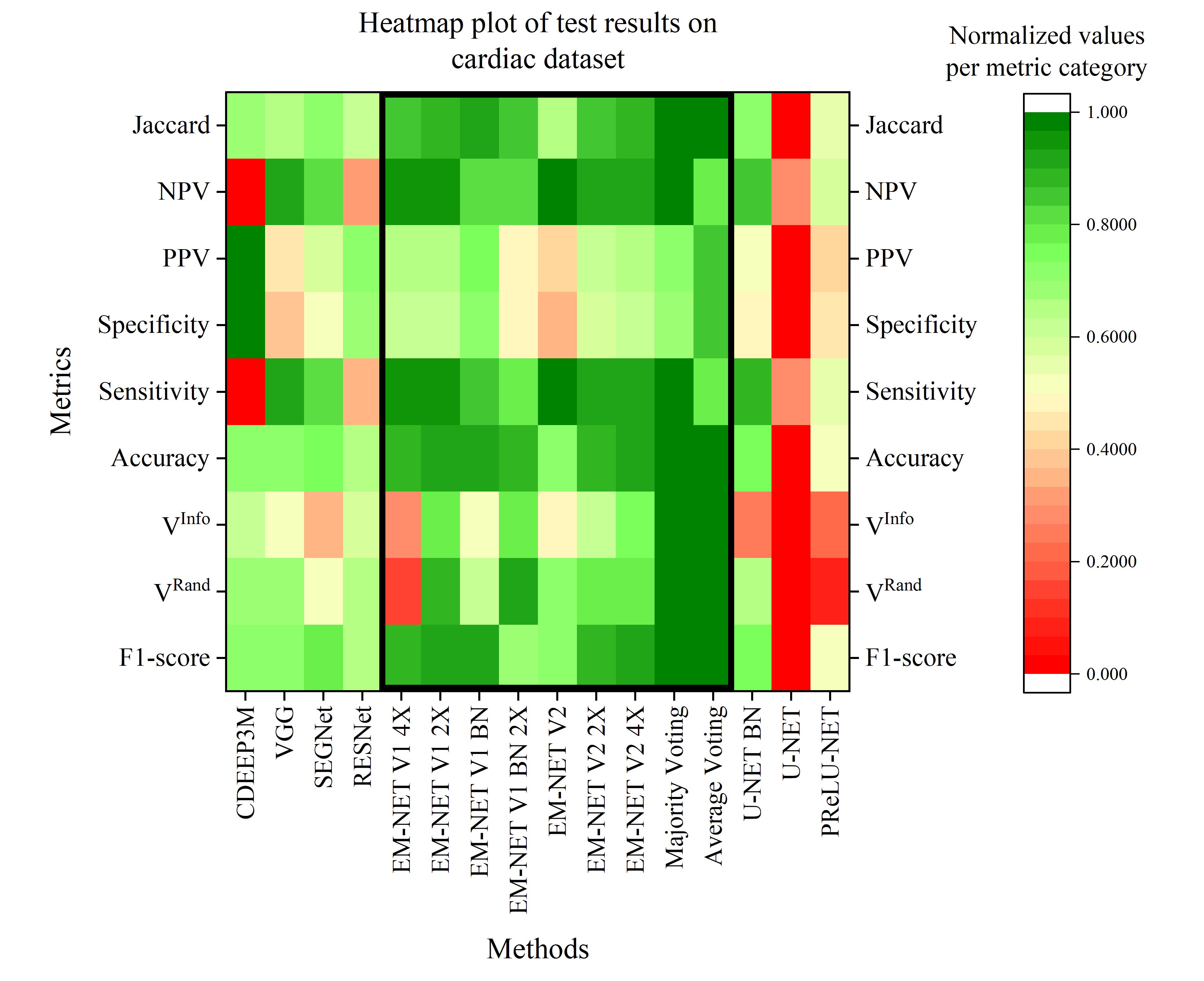 |
Recent high-throughput electron microscopy techniques such as focused ion-beam scanning electron microscopy (FIB-SEM) provide thousands of serial sections which assist the biologists in studying sub-cellular structures at high resolution and large volume. Low contrast of such images hinder image segmentation and 3D visualisation of these datasets. With recent advances in computer vision and deep learning, such datasets can be segmented and reconstructed in 3D with greater ease and speed than with previous approaches. However, these methods still rely on thousands of ground-truth samples for training and electron microscopy datasets require significant amounts of time for carefully curated manual annotations. We address these bottlenecks with EM-net, a scalable deep convolutional neural network for EM image segmentation. We have evaluated EM-net using two datasets, one of which belongs to an ongoing competition on EM stack segmentation since 2012. We show that EM-net variants achieve better performances than current deep learning methods using small- and medium-sized ground-truth datasets. We also show that the ensemble of top EM-net base classifiers outperforms other methods across a wide variety of evaluation metrics.
Related publications:
1 - Khadangi, Afshin, Thomas Boudier, and Vijay Rajagopal. "EM-net: Deep learning for electron microscopy image segmentation.".
|
Automated segmentation of cardiomyocyte Z-disks
Automated segmentation of cardiomyocyte Z-disks from high-throughput scanning electron microscopy data
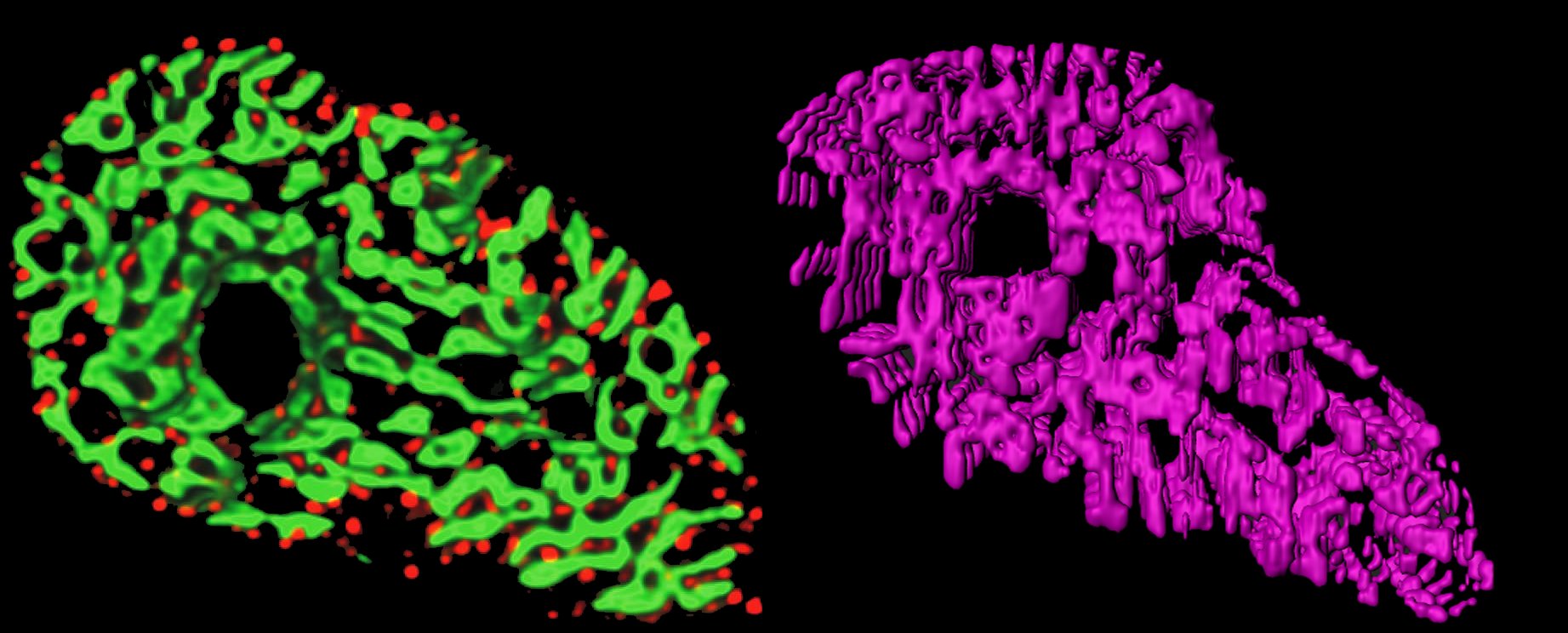 |
In this paper, we proposed to generate a 3D model of Z-disks within single adult cardiac cells from an automated segmentation of a large serial-block-face scanning electron microscopy (SBF-SEM) dataset. The proposed fully automated segmentation scheme is comprised of three main modules including “pre-processing”, “segmentation” and “refinement”. We represent a simple, yet effective model to perform segmentation and refinement steps. Contrast stretching, and Gaussian kernels are used to pre-process the dataset, and well-known “Sobel operators” are used in the segmentation module.
Related publications:
1 - Khadangi, Afshin, Thomas Boudier, and Vijay Rajagopal. "Automated segmentation of cardiomyocyte Z-disks from high-throughput scanning electron microscopy data" BMC Medical Informatics and Decision Making (2019).
|
Automated framework for 3D model reconstruction
Automated framework to reconstruct 3D model of cardiac Z-disk: an image processing approach
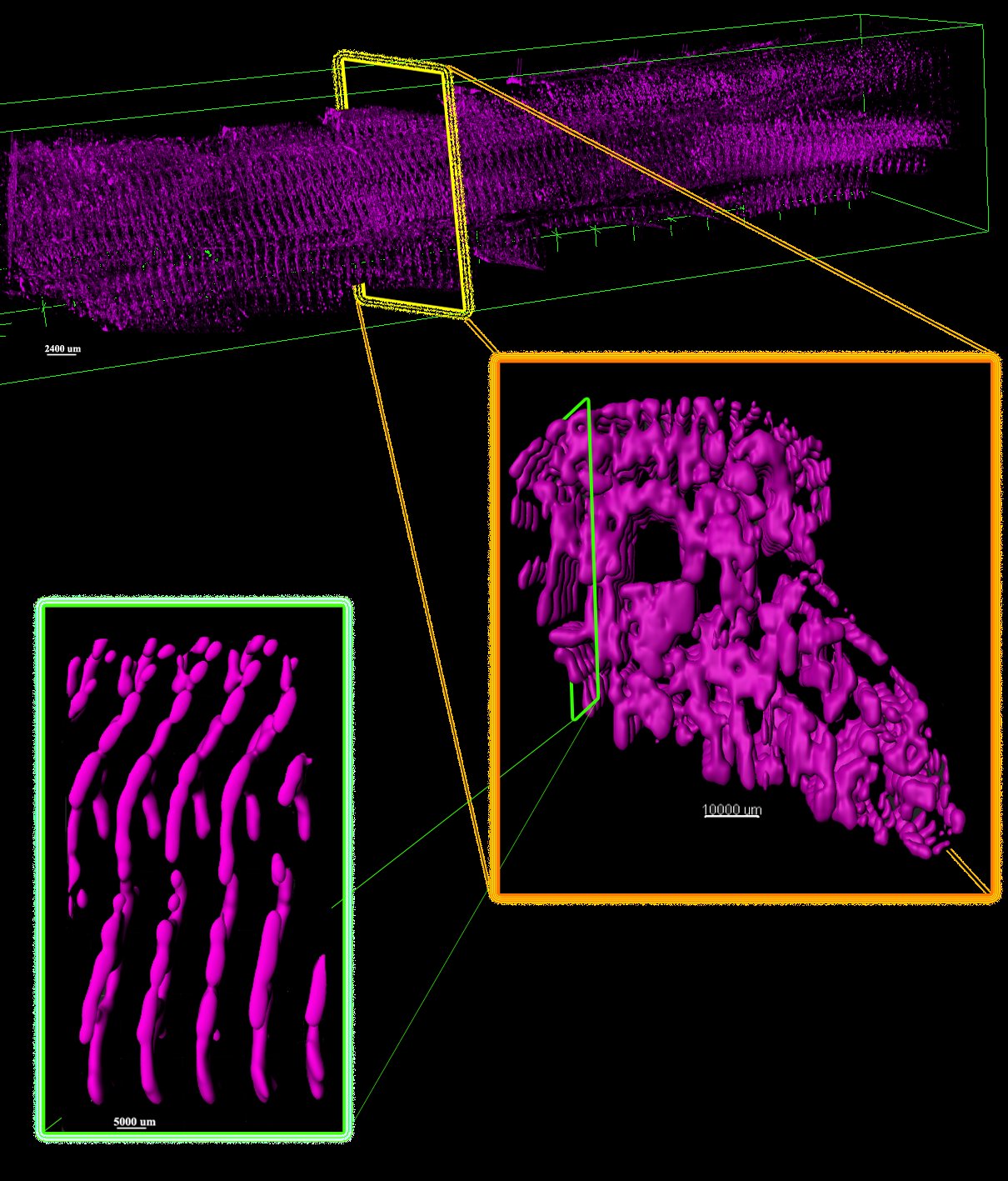 |
The Z-disk or Z-line is located at the lateral borders of sarcomere, the fundamental unit of striated muscle. They provide mechanical stability and can boost contractility of cardiac myocytes. In this paper, we propose to generate a 3D model of Z-disks within single adult cardiac cells from an automated segmentation of a large serial-block-face scanning electron microscopy (SBF-SEM) dataset. The proposed fully automated segmentation scheme is comprised of three main modules including “pre-processing”, “segmentation” and “refinement”. We represent a simple, yet effective model to perform segmentation and refinement steps. Contrast stretching, and Gaussian kernels are used to pre-process the dataset, and well-known “Sobel operators” are used in the segmentation module. We have validated our model by comparing segmentation results with ground-truth annotated Z-disks in terms of pixel-wise accuracy. The results show that our model correctly detects Z-disks with 90.56% accuracy. Finally, the underlying network of Z-disks are rendered in 3D using ImageJ and IMARIS.
Related publications:
1 - Khadangi, Afshin, Thomas Boudier, and Vijay Rajagopal. "Automated framework to reconstruct 3D model of cardiac Z-disk: an image processing approach" 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM).
|
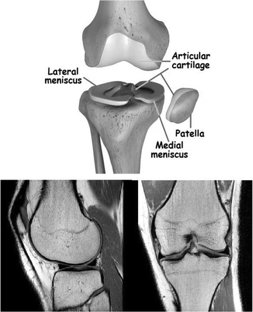
Diagnosis of meniscus tear
A Computer-Aided Type-II Fuzzy Image Processing for Diagnosis of Meniscus Tear
 |
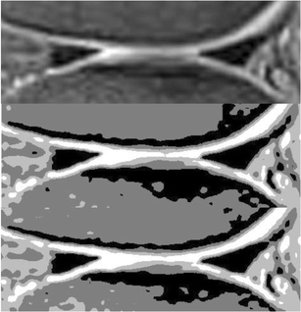 |
Meniscal tear is one of the prevalent knee disorders among young athletes and the aging population, and requires correct diagnosis and surgical intervention, if necessary. Not only the errors followed by human intervention but also the obstacles of manual meniscal tear detection highlight the need for automatic detection techniques. This paper presents a type-2 fuzzy expert system for meniscal tear diagnosis using PD magnetic resonance images (MRI). The scheme of the proposed type-2 fuzzy image processing model is composed of three distinct modules: Pre-processing, Segmentation, and Classification. λ-nhancement algorithm is used to perform the pre-processing step. For the segmentation step, first, Interval Type-2 Fuzzy C-Means (IT2FCM) is applied to the images, outputs of which are then employed by Interval Type-2 Possibilistic C-Means (IT2PCM) to perform post-processes. Second stage concludes with re-estimation of “η” value to enhance IT2PCM. Finally, a Perceptron neural network with two hidden layers is used for Classification stage. The results of the proposed type-2 expert system have been compared with a well-known segmentation algorithm, approving the superiority of the proposed system in meniscal tear recognition.
Related publications:
1 - MHF Zarandi, A Khadangi, F Karimi, IB Turksen. "A Computer-Aided Type-II Fuzzy Image Processing for Diagnosis of Meniscus Tear." Journal of Digital Imaging (2016).
|
Social network analysis
From type-2 fuzzy rate-based neural networks to social networks' behaviors
Social networks like Facebook, Twitter and Instagram are expanding instantly. These rapid changes aggravate the complexity of these networks simultaneously. In this paper, a new approach is proposed to model some complex behaviors of social networks, including “attention inversion”. The model consists of an embedded type-2 fuzzy inference system in collaboration with rate-based neural networks. Finally, an experiment is performed on some selected Twitter hashtags to represent the performance of the model.
Related publications:
1 - A Khadangi, MHF Zarandi "From type-2 fuzzy rate-based neural networks to social networks' behaviors" 2016 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE).
|
Demographics policy analysis
A fuzzy system dynamics approach for evaluation of Iran's new demographic policy
News
August 2021: our preprint CardioVinci: building blocks for virtual cardiac cells using deep learning is available now.
3D renderring of mitochondria (green), myofibrils (red) and Z-disks (blue) using the generated images from GAN
3D renderring of Z-disks using the generated images from GAN
April 2021: our paper EM-stellar: benchmarking deep learning for electron microscopy image segmentation is now published in Bioinformatics
December 2020: EM-stellar got accepted in Bioinformatics
December 2020: My talk is now available online on YouTube
November 2020: I will give a talk at AI Microscopy Symposium hosted by AIVIA and Leica Microsystems
July 2020: our preprint EM-stellar: benchmarking deep learning for electron microscopy image segmentation is available now. This paper has been submitted to Bioinformatics
June 2020: The official implementation of EM-net on Google Colab is now available on YouTube.
June 2020: EM-net will appear on ICPR 2020 proceedings.
May 2020: I have achieved 4th rank globally among more than 800 teams on Google Cloud TPU Kaggle Competition.
April 2020: We have released dEM0ise, a new freely available platform to enhance biological images by removing noise.
April 2020: I have been certified as AWS Developer.
March 2020: I have been certified on Analyzing and Visualizing Data with Microsoft Power BI.
March 2020: I have been certified on End-to-End Machine Learning with TensorFlow on GCP.
February 2020: I have been certified on Amazon SageMaker.
February 2020: our preprint EM-net: Deep learning for electron microscopy image segmentation is available now. This paper has been submitted to ICPR 2020
February 2020: I have been invited as a reviewer panel member of Nature Communications on deep learning.
December 2019: our paper is published on BMC Medical Informatics and Decision Making "Automated segmentation of cardiomyocyte Z-disks from high-throughput scanning electron microscopy data."
July 2019: our proposed method has achieved 1st rank across Australia and 7th globally on ISBI challenge 2012 for Neuronal Stacks Segmentation.
June 2019: I have passed my probation year to continue my Ph.D. as a Ph.D. candidate at the University of Melbourne.
January 2019: We renderred 3D structure of Mitochondria of diabetic cardiac cell relative to the cell volume. The visualization involved processing large volume of data on whole tissue block scale, and not on a single cell level.
January 2019: Our paper previously appeared on IEEE BIBM 2018 proceedings, is selected to be published in the BMC Medical Informatics and Decision Making Special Issue.
December 2018: I gave a talk on segmenting structure of Z-disks from SBF data @IEEE BIBM 2018, held in Madrid, Spain.
November 2018: We renderred 3D structure of Z-disks of control cardiac cell using Serial Block Face data. The results are going to appear on our recent paper.
October 2018: We renderred 3D structure of Mitochondria of diabetic cardiac cell. The visualization involved processing large volume of data on whole tissue block scale, and not on a single cell level.
September 2018: Our paper "Automated framework to reconstruct 3D model of cardiac Z-disk: an image processing approach" is accepted to appear in IEEE BIBM 2018.
August 2018: We are ranked 16 among 199 teams on Kaggle competition to predict whether arbitrary Twitter users follow each other or not. A dataset of 4 million users was processed on TensorFlow.
July 2018: We will collaborate with prof. Thomas Boudier as my new co-supervisor. He is currently bioimage analyst at Centre for Dynamic Imaging of Walter and Eliza Hall Institute of Medical Research.
July 2018: I established a collaborative research work between our group and a leading company based in United States, which is pioneer in the field of high-throughput image analysis and visualization. This partnership will be inaugurated soon.
June 2018: We could segment Z disks of human cardiac myocyte using pre-trained U-nets, via transfer learning.
May 2018: I gave a talk on "Long Short-Term Memory (LSTM) networks" at deep learning journal club.
May 2018: I joined the Deep Learning Journal Club at University of Melbourne.
April 2018: I joined the Department of Biomedical Engineering at University of Melbourne as a Ph.D. student.
CV

My CV
AWARDS AND ACHIEVEMENTS
|
Patents:
Mechanized system for bridge technical management | 2015
A kit for endoscope navigation through the motion of the physician's head | 2014
|
|
Awards:
Melbourne Research Scholarship | The University of Melbourne | 2018
Melbourne School of Engineering Studentship | The University of Melbourne | 2018
Admission to master studies for highly distinguished and talented students | Amirkabir University of Technology | 2013
2nd place in National Iranian Karate Championship (Silver medal) | 2012
1st place in Regional Karate Championship (Gold medal) | 2012
|
|
Achievements:
dEM0ise: zero cost dynamic deep learning for zero-noise microscopy data |free online platform, image enhancement (www.dem0ise.com) |2020
1st place in Australia and 8th place worldwide | Massachusetts Institute of Technology ISBI challenge Neuronal Stacks Segmentation (Challenge leaderboard) |2020
Selection of the conference paper as special issue publication | 2019
5th place in first Idea Bazaar for Biomedical Engineering among more than 61 ideas nationwide | Amirkabir University of Technology | 2013
Most young achiveving teaching assistantship | Amirkabir University of Technology | 2010
Placed 578 among more than 300,000 students participating nationwide entrance exam for undergraduate studies | 2009
|
Accomplishments
|
Publications:
Journal papers:
Khadangi, Afshin, Eric Hanssen, and Vijay Rajagopal. "Automated segmentation of cardiomyocyte Z-disks from high-throughput scanning electron microscopy data." BMC Medical Informatics and Decision Making 19, no. 6 (2019): 1-14.
Zarandi, MH Fazel, A. Khadangi, F. Karimi, and I. B. Turksen. "A computer-aided type-II fuzzy image processing for diagnosis of meniscus tear." Journal of digital imaging 29, no. 6 (2016): 677-695.
Conference papers:
Khadangi, Afshin, Eric Hanssen, and Vijay Rajagopal. "Automated framework to reconstruct 3D model of cardiac Z-disk: an image processing approach." In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 877-884. IEEE, 2018.
Khadangi, A., and MH Fazel Zarandi. "From type-2 fuzzy rate-based neural networks to social networks' behaviors." In 2016 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), pp. 1970-1975. IEEE, 2016.
Khadangi, A., A. Ghahari, and MH Fazel Zarandi. "A fuzzy system dynamics approach for evaluation of Iran's new demographic policy." In 2015 Annual Conference of the North American Fuzzy Information Processing Society (NAFIPS) held jointly with 2015 5th World Conference on Soft Computing (WConSC), pp. 1-6. IEEE, 2015.
Preprint:
Khadangi, Afshin, Thomas Boudier, and Vijay Rajagopal. "EM-net: Deep learning for electron microscopy image segmentation." bioRxiv (2020).
|
|
Organisations and memberships:
Nature Research| Jan 2020 – Present
Invited reviewer on Deep Learning
Iran’s National Elite Foundation | Feb 2015 – Present
Distinguished young inventor and talented student
Institute of Electrical and Electronics Engineers (IEEE) | 2016 – 2018
Invited reviewer on machine learning and image analysis for FUZZ-IEEE
|
|
Licenses and Certifications:
AWS Developer | Amazon Web Services (AWS)| 2020
End-to-End Machine Learning with TensorFlow on GCP |Coursera|2020
Analyzing and Visualizing Data with Microsoft Power BI | Microscoft | 2020
2020 AWS SageMaker, AI and Machine Learning | Udemy | 2020
Java Programming Masterclass for Software Developers | Udemy | 2020
|
|
Teaching assistantship:
MATLAB Programming | University of Melbourne | March 2020 – present
Machine Learning | Amirkabir University of Technology | 2014 – 2015
Materials Science | Amirkabir University of Technology | 2010 – 2012
|
Education
|
Doctor of Philosophy | The University of Melbourne | 2018 – 2021 (EXPECTED)
Biomedical Engineering
Thesis title: Machine Learning approaches to generate computational models of the cell
Master of Science | Amirkabir University of Technology | 2013 – 2015
Industrial Engineering – Healthcare Systems Engineering
Thesis title: Type-2 Fuzzy Expert System for Prostate Cancer Grading
Bachelor of Science | Amirkabir University of Technology | 2009 – 2013
Industrial Engineering – Systems Engineering
Thesis title: Fuzzy Expert System for Prostate Gland Boundary Delineation
|
Contact
Elements
Text
This is bold and this is strong. This is italic and this is emphasized.
This is superscript text and this is subscript text.
This is underlined and this is code: for (;;) { ... }. Finally, this is a link.
Heading Level 2
Heading Level 3
Heading Level 4
Heading Level 5
Heading Level 6
Blockquote
Fringilla nisl. Donec accumsan interdum nisi, quis tincidunt felis sagittis eget tempus euismod. Vestibulum ante ipsum primis in faucibus vestibulum. Blandit adipiscing eu felis iaculis volutpat ac adipiscing accumsan faucibus. Vestibulum ante ipsum primis in faucibus lorem ipsum dolor sit amet nullam adipiscing eu felis.
Preformatted
i = 0;
while (!deck.isInOrder()) {
print 'Iteration ' + i;
deck.shuffle();
i++;
}
print 'It took ' + i + ' iterations to sort the deck.';
Lists
Unordered
- Dolor pulvinar etiam.
- Sagittis adipiscing.
- Felis enim feugiat.
Alternate
- Dolor pulvinar etiam.
- Sagittis adipiscing.
- Felis enim feugiat.
Ordered
- Dolor pulvinar etiam.
- Etiam vel felis viverra.
- Felis enim feugiat.
- Dolor pulvinar etiam.
- Etiam vel felis lorem.
- Felis enim et feugiat.
Icons
Actions
Table
Default
| Name |
Description |
Price |
| Item One |
Ante turpis integer aliquet porttitor. |
29.99 |
| Item Two |
Vis ac commodo adipiscing arcu aliquet. |
19.99 |
| Item Three |
Morbi faucibus arcu accumsan lorem. |
29.99 |
| Item Four |
Vitae integer tempus condimentum. |
19.99 |
| Item Five |
Ante turpis integer aliquet porttitor. |
29.99 |
|
100.00 |
Alternate
| Name |
Description |
Price |
| Item One |
Ante turpis integer aliquet porttitor. |
29.99 |
| Item Two |
Vis ac commodo adipiscing arcu aliquet. |
19.99 |
| Item Three |
Morbi faucibus arcu accumsan lorem. |
29.99 |
| Item Four |
Vitae integer tempus condimentum. |
19.99 |
| Item Five |
Ante turpis integer aliquet porttitor. |
29.99 |
|
100.00 |












